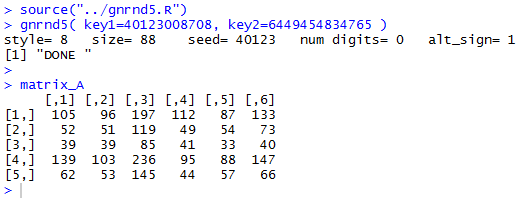
source("../gnrnd5.R")
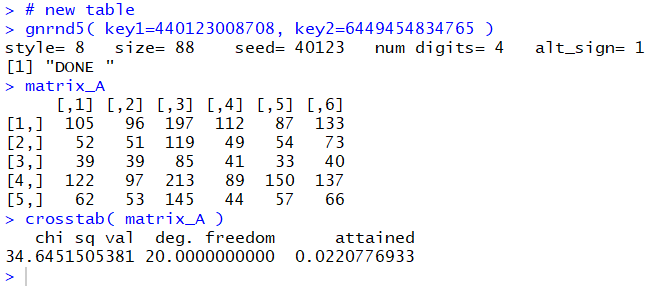
gnrnd5( key1=40123008708, key2=6449454834765 )
matrix_A
as is shown in Figure 1.

crosstab()
has been designed to perform all of those steps. In fact,
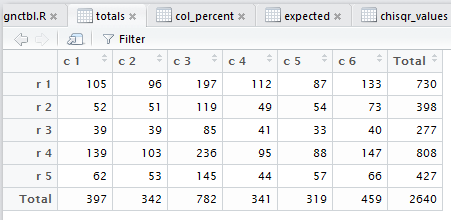
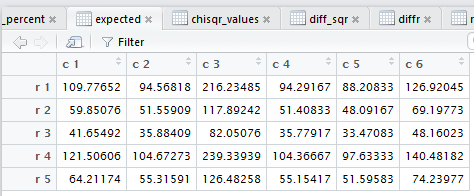
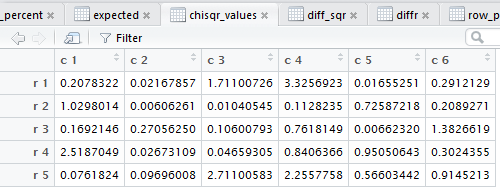
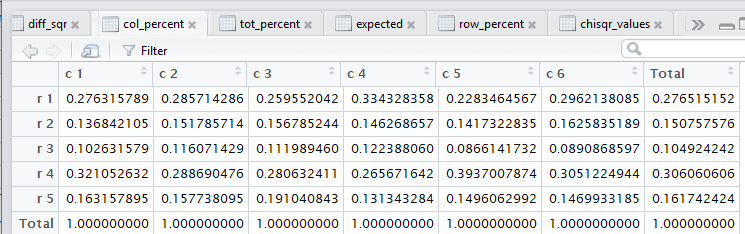
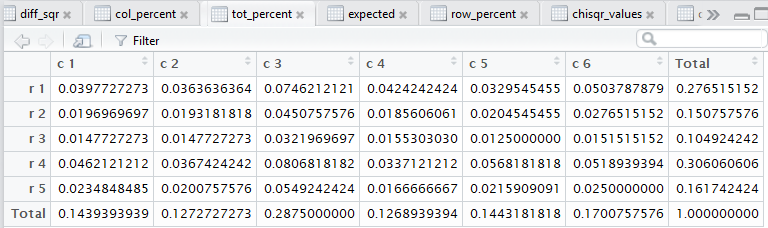
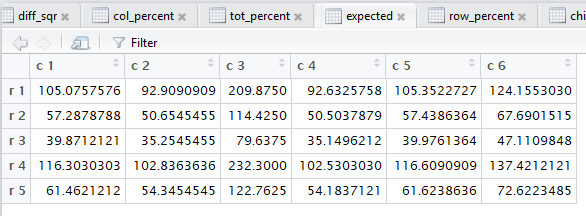
crosstab() also computes the
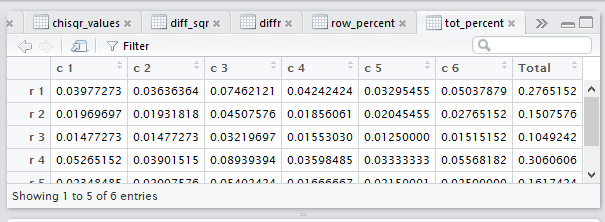
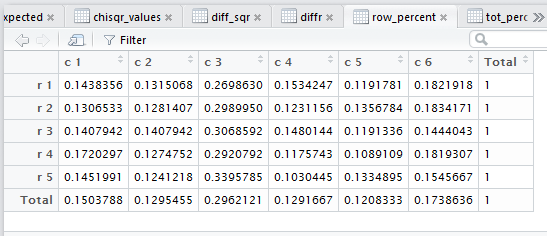
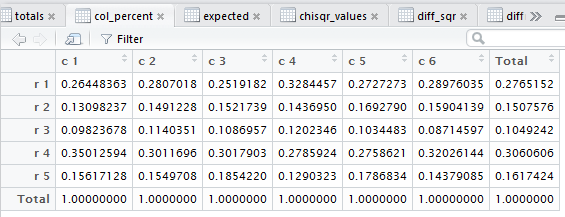
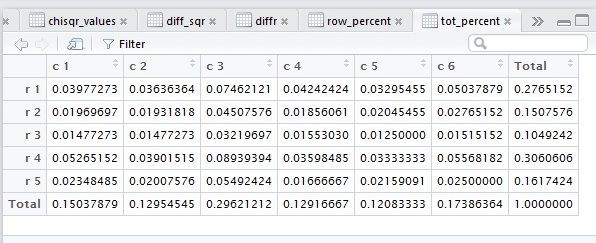
row percents, the column percents, and the total percent
for the data. Beyond that, also as noted in that earlier page,
crosstab()
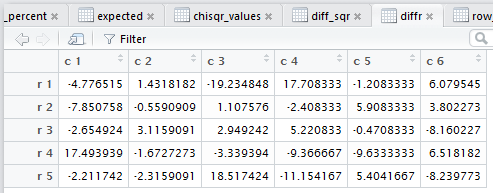
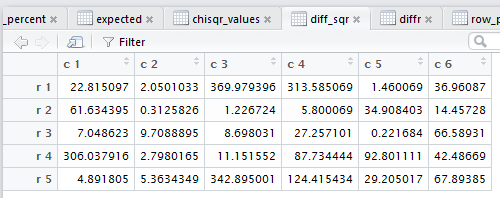
goes through and displays the tabular results for each of the steps
needed to do the χ² computation.
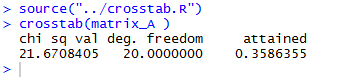
crosstab() function. The R commands:
source("../crosstab.R")
crosstab(matrix_A )
produced the console output shown in
Figure 2.
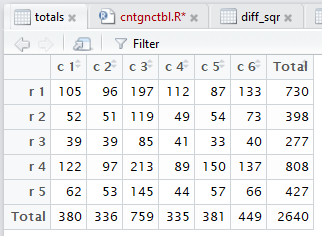
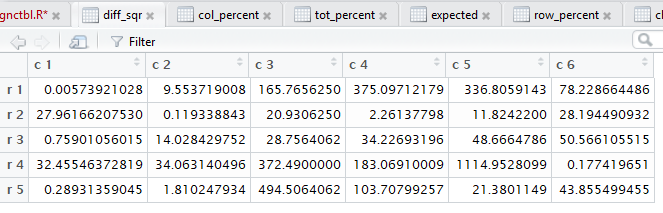
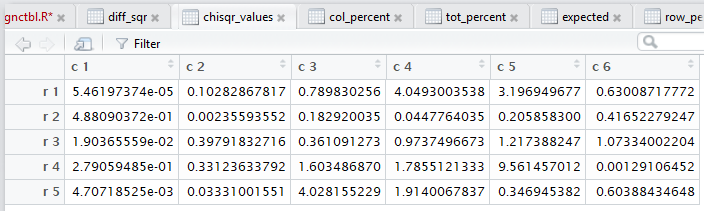
crosstab() produces,
and then displays in the editor pane,
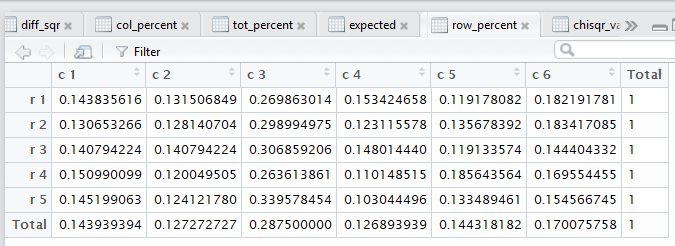
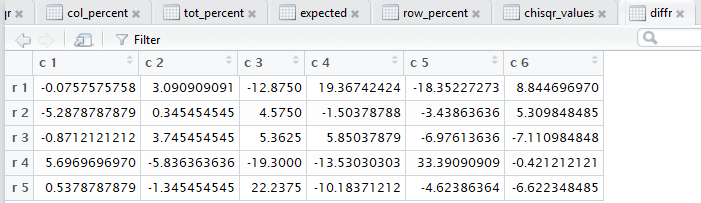
all of those steps. Figure 3 captures that editor pane from my
computer. In Figure 3 we see that there are now many tabs
for different windows in that pane. It may be hard to see because of
the lack of significant contrast, but the tot_percent
tab is highlighted meaning that the display is showing
that set of values.