# the commands used on frimgrouped.htm
# Create the list of frequencies
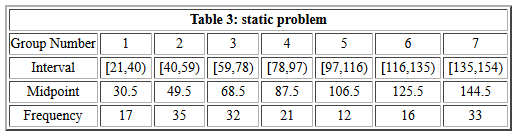
freq <- c(17,35,32,21,12,16,33)
freq
# Create the midpoint values. Note that there are
# many different ways to generate these values.
# Here we will just enter them as a list.
mp<-c(30.5,49.5,68.5,87.5,106.5,125.5,144.5)
mp
# Just to demonstrate another approach we will
# use the seq() function
alt_mp <- seq(from=30.5,to=144.5,by=19)
alt_mp
#
# Now create a list that holds each of the
# midpoint values the number of times given
# by the corresponding frequency value
x<-rep(mp,freq)
x
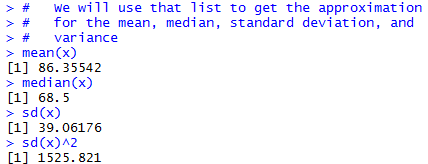
# We will use that list to get the approximation
# for the mean, median, standard deviation, and
# variance
mean(x)
median(x)
sd(x)
sd(x)^2
#
# The work above gave our best approximation given
# the table that we had. Let us look at a contrived
# counter example. We use the same frequencies but
# how about using values other than the midpoints.
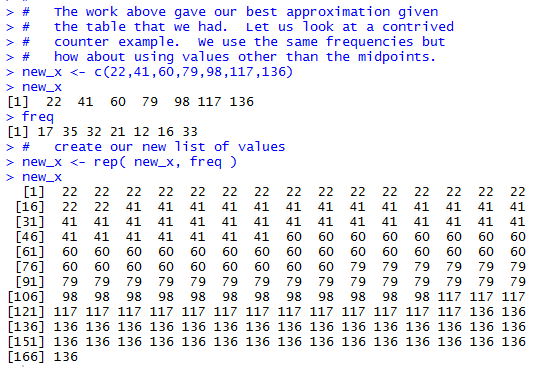
new_x <- c(22,41,60,79,98,117,136)
new_x
freq
# create our new list of values
new_x <- rep( new_x, freq )
new_x
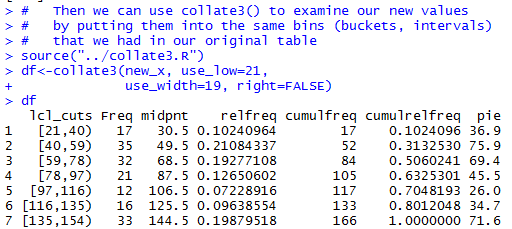
# Then we can use collate3() to examine our new values
# by putting them into the same bins (buckets, intervals)
# that we had in our original table
source("../collate3.R")
df<-collate3(new_x, use_low=21,
use_width=19, right=FALSE)
df
# The frequency tables is exactly that of our original
# table. Now look at the mean, median, abd
# standard deviation
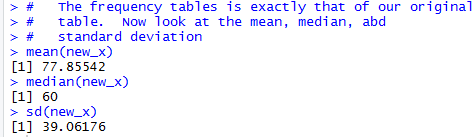
mean(new_x)
median(new_x)
sd(new_x)
#
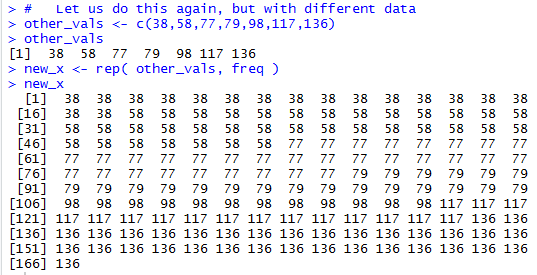
# Let us do this again, but with different data
other_vals <- c(38,58,77,79,98,117,136)
other_vals
new_x <- rep( other_vals, freq )
new_x
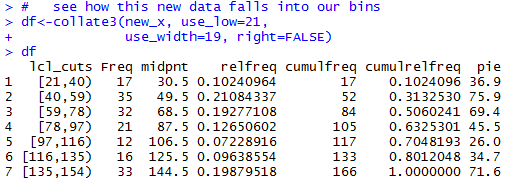
# see how this new data falls into our bins
df<-collate3(new_x, use_low=21,
use_width=19, right=FALSE)
df
# Again, these intervals are just like our original
# table. Now look at the mean, median, abd
# standard deviation
mean(new_x)
median(new_x)
sd(new_x)
#